|
RUSSIAN ACADEMY OF SCIENCES Shemyakin & Ovchinikov Institute of Bioorganic Chemistry | |

| |
| | Home | People | Research | Publications | Protocols | Collaborations | |
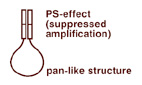
Publications |
Selective suppression of the polymerase chain reaction and methods based upon itLukyanov S, Lukyanov K, Tarabykin V, Fradkov A, Rebrikov D, Matz M, Britanova O, Shagin D, Bogdanova E, Gurskaya N, Vagner L, Usman N. The most important processes in various biological systems (cell differentiation and morphogenesis during embryonic development and regeneration, apoptosis or malignization of cells, etc.) are under the control of specific regulatory genes. To understand the underlying molecular mechanisms, the genes involved in their regulation should be revealed and studied. At present, the majority of methods of molecular biology involved in unraveling such problems are based on PCR. However, the use of PCR requires information on the sequence of the DNA under study. When the sequence is partially or completely unknown, the PCR often encounters difficulties. The PCR suppression effect, discovered in our laboratory, generated a number of Highly effective, mutually complementary methods of finding and analyzing new functionally important DNA and RNA sequences when information, complete or partial, on their primary structure is absent. The use of suppression PCR enables one to skip labor-intensive and not very effective methods of physical fractionation of DNA and makes the methods based upon it easier, more rapid and reproducible. The methods developed on Suppression PCR effect (cDNA library construction starting from small amount of total RNA, Marathon cDNA RACE, Suppression Subtractive Hybridization, Ordered Differential Display, genome walking, in vitro cloning and others) display a high effectiveness and, taken together, enable intricate DNA analyses to be performed – from the search for new sequences to the total sequencing of the corresponding genes. |