Подразделение было расформировано и с 2023 года входит в состав Лаборатория оптического биоимиджинга.
Группа молекулярных меток для оптической наноскопии
|
Руководитель: Мишин Александр Сергеевич |
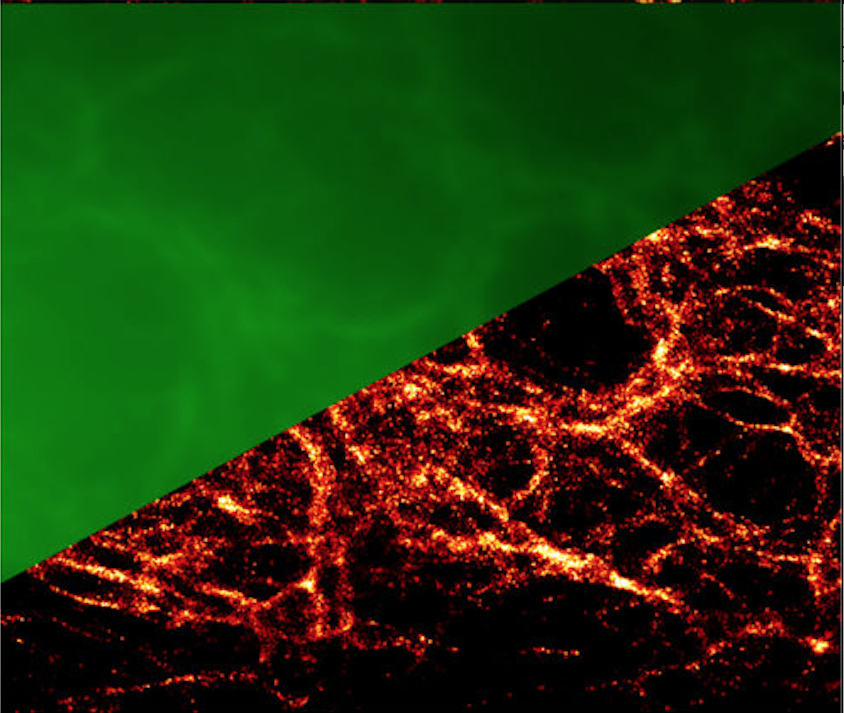
В Группе ведутся работы по созданию репортерных систем, адаптированных для использования в прижизненной наноскопии. Группа образована в 2018 г.
- метод Protein-PAINT: обратимое взаимодействие генетически кодируемого белка-репортера и низкомолекулярного красителя, свободно проникающего через цитоплазматическую мембрану, позволяет достигнуть высокой фотостабильности и плотности мечения целевых структур. Созданы зеленые, оранжевые, а также красные пары флуороген-белок, подходящие для методов локализационной микроскопии (SMLM) а также STED
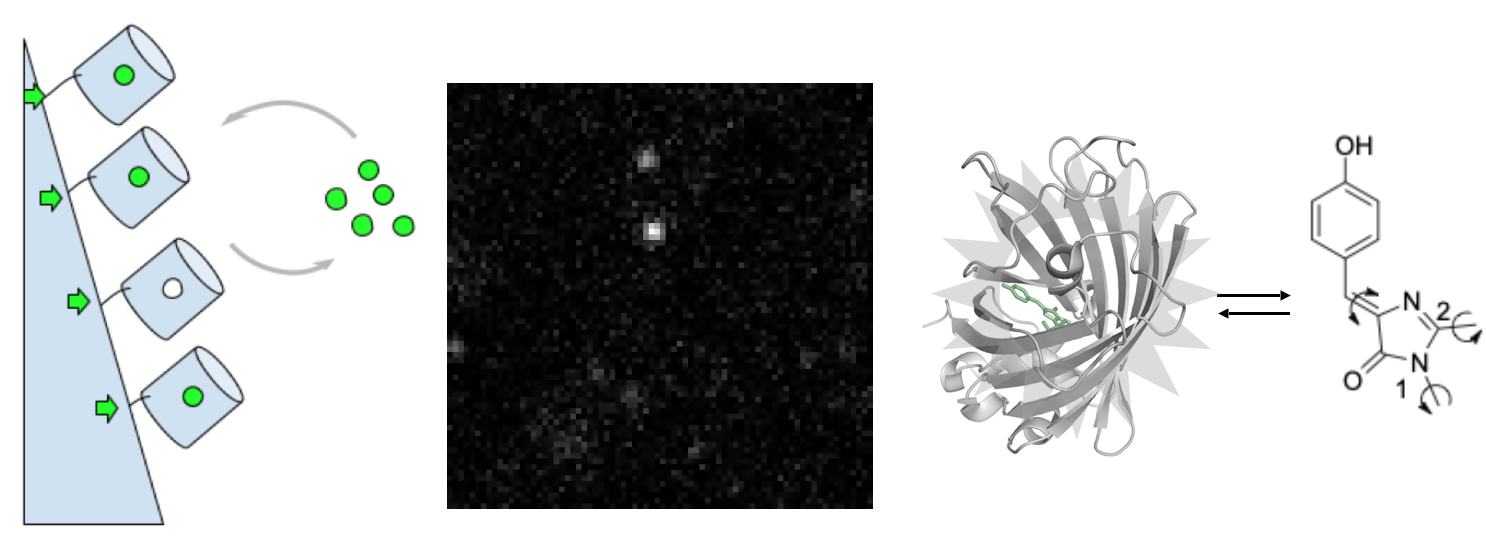
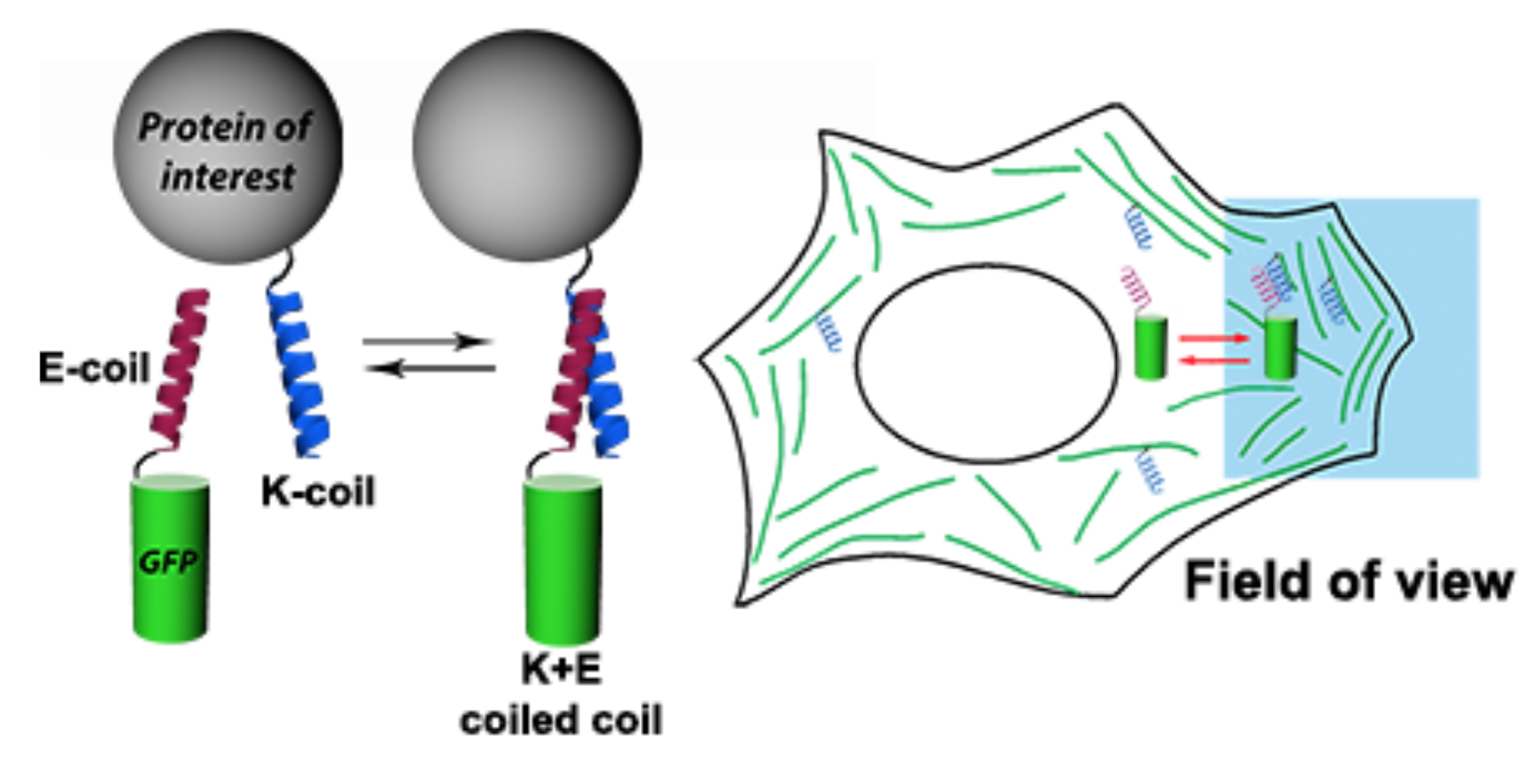
- метод Protein-PAINT / IRES: полностью генетически кодируемая версия protein-PAINT, основанная на обратимом взаимодействии коротких пептидов (K/E-спиралей). Созданная система увеличивает плотность мечения целевых структур при наноскопии в живых клетках
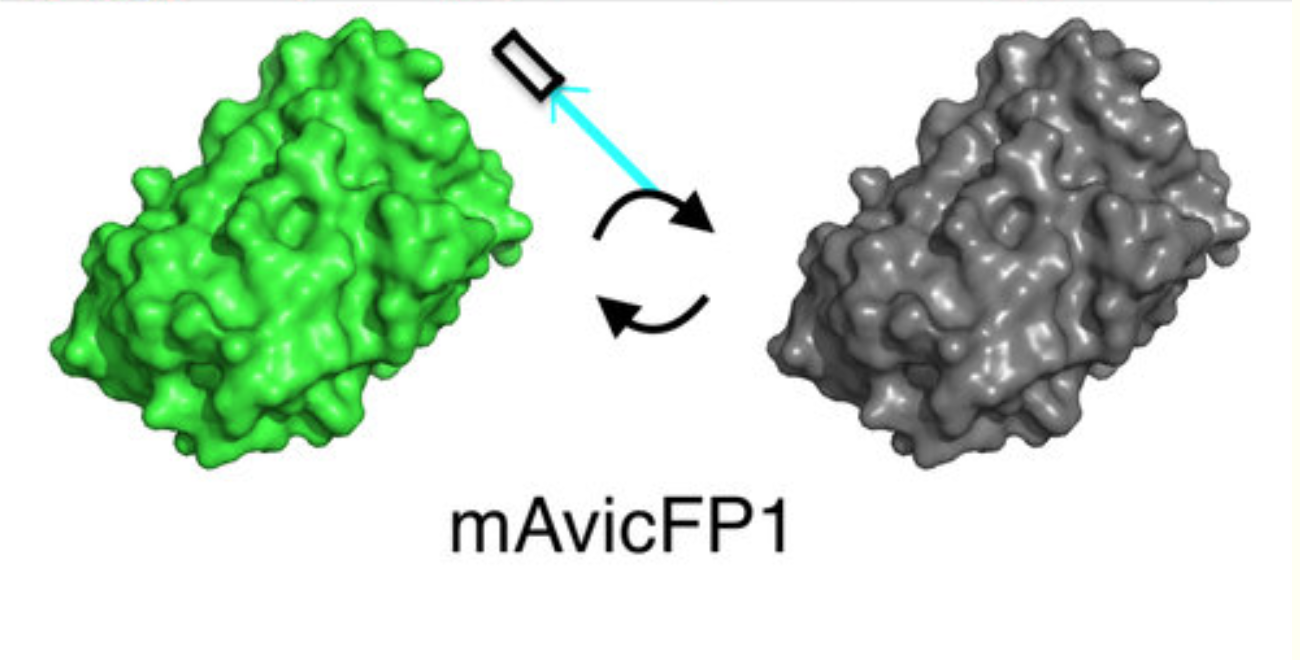
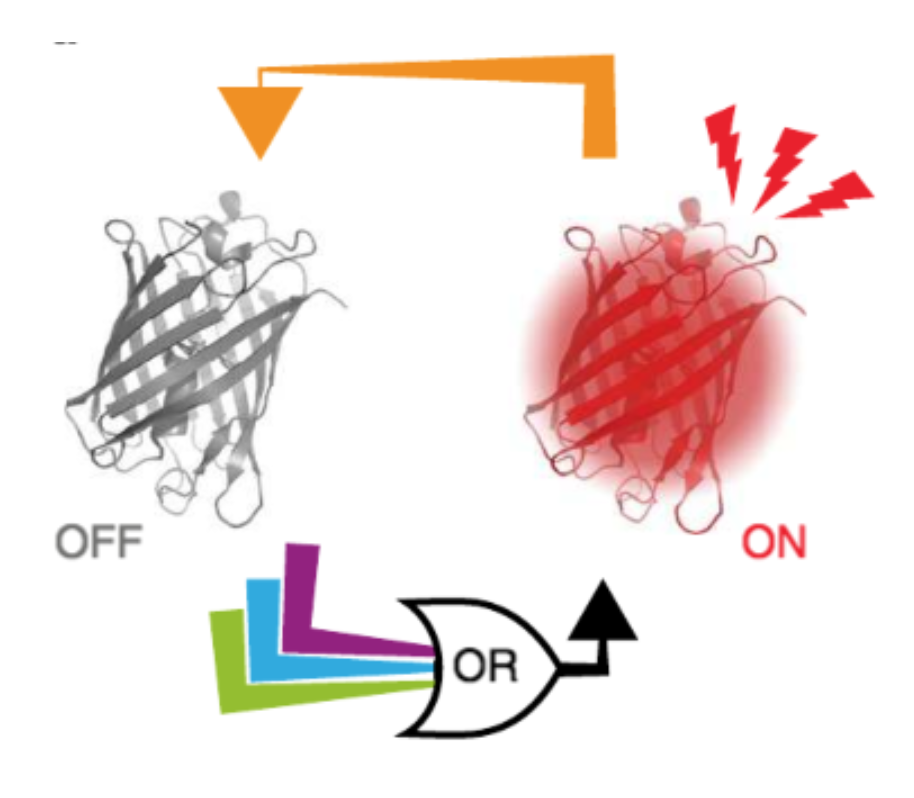
- спонтанно мигающие флуоресцентные белки и красители
- метки для STED и RESOLFT
- люминисцентная микроскопия и имиджинг
| ФИО | Должность | Контакты |
|---|
 Загрузка...
Загрузка...Научные проекты
 Загрузка...
Загрузка...Мишин Александр Сергеевич
Москва, ул. Миклухо-Маклая, 16/10 — На карте
 Загрузка...
Загрузка...
