Press-room / news / Science news /
Start codon context and occurrence of AUG codons in the beginning of protein coding open reading frames co-evolve
Researchers from IBCh RAS together with their international collаborators discovered an evolutionary link between efficiency of start codons and the probability of AUG codon occurrence in the 5'-end parts of protein-coding sequences. When start codons are inefficient the next AUG codon is more likely to be found in the same reading frame. At the same time in case of highly efficient start codons, the next AUG codon is more likely to be found in alternative frames. They also have shown that weak initiation at the first starts is associated with the synthesis of shortened proteoforms as a result of initiation at the second starts.
According to our current understanding of the translation initiation in eukaryotes, the small subunit of the ribosome with initiation factors binds to the 5’ end of the mRNA, and then begins to scan the mRNA in search for an initiator codon. Once start codon (in most cases this is AUG) is found, a chain of events occurs, leading to the dissociation of initiating factors, a large subunit joining and the beginning of the synthesis of the polypeptide. The efficiency of start codon recognition depends on its closest nucleotide context. In the 1980s, Marilyn Kozak showed that the optimal context depends primarily on nucleotides at positions -3 and +4 (A in the AUG is +1). Since then the immediate nucleotide context for initiation is usually referred to as Kozak context. If the context is not optimal, then a large proportion of scanning ribosomes can skip it and continue scanning further in the process that is known as the leaky scanning.
Researchers from the IBCh RAS together with colleagues from Moscow State University, University of Granada in Spain and from University College Cork in Ireland investigated the link between Kozak context and occurrence of AUG codons immediately downstream. When the start codon of the protein coding open reading frame is not in the optimal context, there are two possibilities for “leaky” ribosomes. If the next starting codon is in the same reading frame, then as a result of its recognition, the ribosome will synthesize a shortened version of the protein. If the next starting codon is in one of the two other reading frames, a completely different polypeptide will be synthesized. It is unlikely that such an initiation product may have any functional role in the cell.
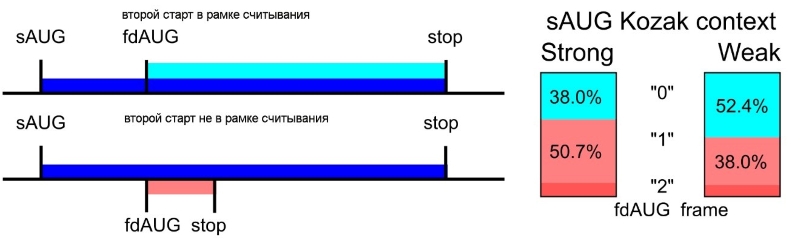
Left: Two configurations of first AUG codons in protein-coding sequences. Right: Frequency of such configurations for start codons of different strengths.
The researchers investigated the relative positions of the first (main) and subsequent AUG codons in human mRNAs and found that the probability of finding the second codon in different reading frames depends on the context of the first codon. The weaker the initiating context of the first AUG codon, the more likely the second AUG codon is in the same main reading frame. Moreover, comparative analysis of the contexts of the start codons across the genomes of different vertebrate indicates that the low efficiency of the first start is associated with the high efficiency of the second start (when in the same reading frame). In other words, the mRNA architecture, that allows two protein products to be synthesized from one mRNA due to leaky scanning is under evolutionary selection in some genes. The mechanism of leaky scanning allows diversification of the proteome by producing proteins with different N-termini from the same mRNA. It is very likely that such altered proteoforms may have alternative intracellular localization, or even functional activity.
The study was published in the journal Genome Research.
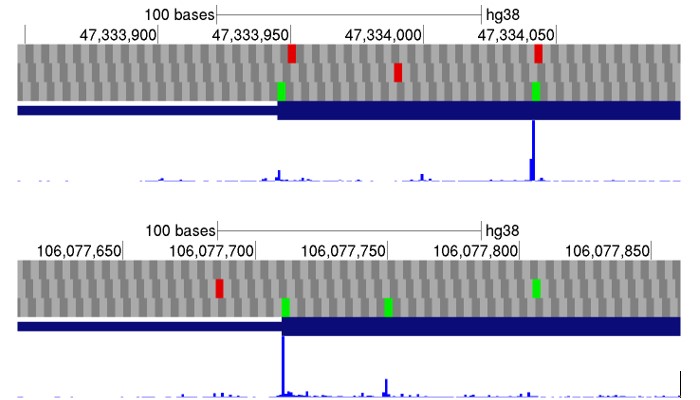
Examples of two start codon utilizations in human genes (AUG are in green) supported by ribosome profiling data (blue peaks correspond to ribosomes inhibited during initiation) available in GWIPS-viz browser (https://gwips.ucc.ie)
july 17, 2020




