Press-room / news / Science news /
Evolution of Phage Tail Sheath Protein
A team of scientists from the Laboratory of molecular bioengineering IBCh RAS analysed 112 contractile phage tail sheath proteins (TShP) representing different groups of bacteriophages and archaeal viruses with myoviral morphology have been modelled with the novel machine learning software, AlphaFold 2. The common core domain of all studied sheath proteins, including viral and T6SS proteins, comprised both N-terminal and C-terminal parts, whereas the other parts consisted of one or several moderately conserved domains, presumably added during phage evolution.
The analysis of experimentally determined and modelled structures indicates the presence of a common conserved core inherent for all analysed sheath proteins, including phage tail sheath proteins (TShPs), type VI secretion system sheath proteins, bacteriocin, and anti-feeding prophage sheath proteins. Most of the phage TShPs studied in this report differed from non-phage sheath proteins due to the presence of additional domains. These domains seem to be located away from the part of the TShP that is in contact with the tail tube. Such a location would prevent perturbation of the tail assembly and the function of the contractile mechanism. In this way, the evolution of phage TShPs was conditioned by its biological role. Phylogenetic analysis on the basis of structural similarity indicated the relatedness of T6SS sheath proteins and the TShP of giant phages. This may indicate the ancient divergence of phage sheath proteins and T6SS. Further, the similarity between anti-feeding prophage sheath proteins, the TShPs of Gram-positive bacteria, and the predicted sheath proteins for archaea may also have an ancient origin and were a consequence of the specialisation of AFP. In contrast, diffocin and pyocin sheath proteins appear to arise later and can be polyphyletic. As a minimum, the Peptoclostridium difficile diffocin sheath protein is structurally closer to Clostridium phages than it is to all other sheath proteins, and the Pseudomonas aeruginosa pyocin sheath protein is structurally similar to the TShPs of phages infecting beta- and gammaproteobacteria. For the future, the origin of phage-like contractile machines will require dedicated research using a larger representative group.
The size of the phage tail sheath proteins and the number of additional domains correlated with the size of the genome, such that small phages possessed shorter one- or two-domain TShPs. This observation seems reasonable, since the additional domains are not essential for the assembly and operation of the contractile mechanism, but they will consume resources for carrying the extra genetic material and protein synthesis during the infection. Large phages often have multi-domain TShPs. The suggestion, here, is that the ancestral form of phage TShPs possessed one main domain, and that during the evolution of phages, accompanied by an increase in genome size, phage TShPs acquired additional domains. This process may have some common features with the acquisition of additional functional genes during Jumbo phage evolution.
The necessity of expending additional resources as a result of having larger sheath proteins must be justified by competitive advantages provided by the additional domains. Most additional domains in the studied TShPs exhibited an immunoglobulin-like fold. It might be hypothesised that the presence of additional Ig-like TShP domains assists the adhesion of phages to the bacteria. Ig-like domains have been shown to be the subject of common horizontal exchange between diverse classes of both lytic and temperate phages, and Ig-like domains may play an accessory role in phage infection by weakly interacting with carbohydrates on the bacterial cell surface. A further hypothesis might be that these domains can participate in the formation of tail appendages detected for some large phages, since Ig-like domains often participate in protein-protein interactions, which, in turn, also promote cell adhesion. It might also be possible that the presence of additional domains facilitates an increase in the stability of the virion, which is vital for phages, by cementing the assembled tail exploiting the interactions between additional domains of TShPs.
The results are published in the Viruses.
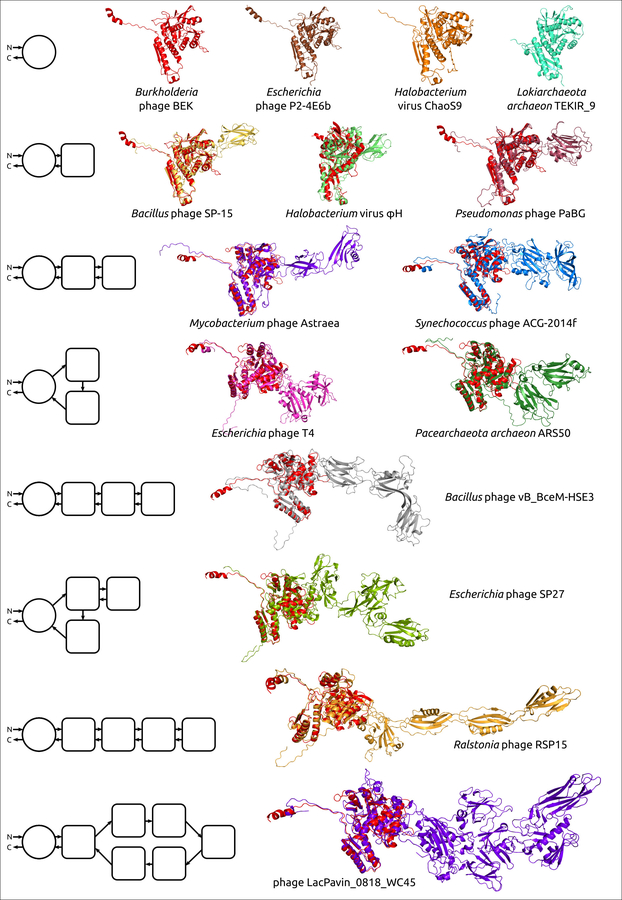
Figure 1. Examples of the structural architecture for the modelled contractile phage sheath proteins. The TShPs consisting of two and more domains are superimposed with the modelled structure of Burkholderia phage BEK tail sheath protein, depicted in the red colour. The schemes on the left show the structural architecture of proteins. The main domain is depicted as a circle, and the additional domains are represented as squares with rounded corners. The direction of the polypeptide chain from the N- to C-termini is shown with arrows.
june 16, 2022


