Press-room / news / Science news /
Capsule-Targeting Depolymerases Derived from Acinetobacter baumannii Prophage Regions
A team of scientists from the Laboratory of molecular bioengineering IBCh RAS together with the colleagues from other Russian Institutes bioinformatically predicted and recombinantly produced several different depolymerases encoded in the prophage regions of Acinetobacter baumannii genomes. For two depolymerases, the specificity to capsular polysaccharides (CPSs) of A. baumannii belonging to K1 and K92 capsular types (K types) was determined. These enzymes can be considered as suitable candidates for the development of new antibacterials against corresponding A. baumannii K types.
Six structural depolymerases encoded in different A. baumannii genomes were bioinformatically predicted, recombinantly produced, and studied. The examination of bacterial genomes, where the sequences were found, testified that they apparently belonged to prophage regions containing genes encoding phage assembly and structural proteins, products involved in phage nucleotide metabolism, the packaging of DNA into the capsid, bacterial cell lysis, and integration of phage DNA into bacterial genomes.
Protein remote homology detection by HMM-HMM comparison conducted with HHpred demonstrated that the depolymerases possessed a multi-domain structure. The N-terminal parts of some depolymerases shared a similarity with the Escherichia phage T7 tail fiber. The remaining parts of all the proteins analysed contained the regions that showed similarities with different carbohydrate-hydrolyzing enzymes or lytic phage tailspikes. Thus, the studied proteins most likely assigned to prophage tailspike depolymerases, which participate in the first steps of interactions with bacterial hosts. Prediction of the domain organization of prophage-derived depolymerase monomers with AlphaFold 2 also revealed several regions corresponding to the N-terminal, central, and C-terminal parts.
Deletion mutants lacking the N-terminal domains responsible for the attachment of capsular polysaccharides (CPSs) recognizing/degrading parts of the prophage-derived depolymerases to the phage particles were cloned, expressed, and purified. The strategy of cloning only receptor-binding/recognizing parts of the depolymerases was chosen in order to avoid possible protein aggregation due to the hydrophobicity of their N-termini. The specificity of purified prophage-derived depolymerases was tested using a collection of A. baumannii strains belonging to 56 different capsular types. Two of six recombinant proteins expressed in E. coli, namely DpoAB5075 and DpoB8300, exhibited depolymerase activities on the bacterial lawns of A. baumannii AYE and B8300 belonging to K1 and K92 capsular types, respectively.
Taking into account that the CPSs are the primary receptors for depolymerase-carrying lytic A. baumannii phages, the first step of the infection of bacterial host by a temperate phage can also cause the degradation of corresponding CPS with a specific structural depolymerase. This is confirmed by the fact that prophage-derived depolymerase DpoB8300 degrades CPS of the K92 capsular type, to which the host strain A. baumannii B8300 also belongs. However, A. baumannii AB5075-UW, a representative of the K25 capsular type, encoded depolymerase DpoAB5075 which possess the enzymatic activity towards K1 CPS. This could be explained by possible recombination events between bacterial genomes or the horizontal transfer of prophage-containing regions. The analysis of oligosaccharide products obtained by the degradation of the A. baumannii CPSs by recombinant prophage-derived depolymerases DpoAB5075 and DpoB8300 showed that the enzymes were specific glycosidases that cleaved the CPSs by the hydrolytic mechanism to produce a monomer and oligomers of the K1 and K92 units, respectively.
The capsule comprising repeating polysaccharide units is a major A. baumannii virulence determinant, which protects bacteria from host immune defences. Thus, CPS-degrading enzymes or depolymerases encoded in phage genomes can represent effective antivirulence agents. The antivirulence efficacy of several depolymerases encoded in the genomes of lytic A. baumannii-phages was explored using a Galleria mellonella model. In particular, a specific K2 depolymerase (B3gp42) encoding in the phage vB_AbaP_B3 genome (GenBank: MF033348) was shown to protect larvae from bacterial infections, using either pretreatments or single-enzyme injections after bacterial challenges in a dose-dependent manner. In another study, depolymerase Dpo48 encoded in phage IME200 genome (GenBank: KT804908) was shown to reduce the virulence of A. baumannii host cells.
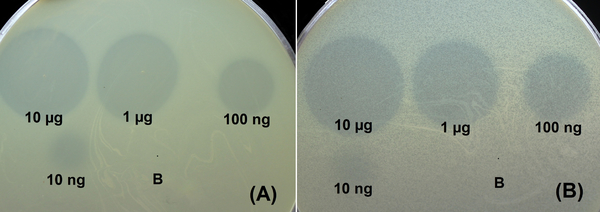
Figure 1. Spot test with serial 10-fold titration of purified recombinant depolymerases DpoB8300 (A) and DpoAB5075 (B) and on A. baumannii B8300 and AYE lawns, respectively, after 18 h of incubation; B-buffer (20 mM Tris-HCl, pH 7.5, and 150 mM NaCl) for storage of the proteins as a negative control.
In this study, for the first time, we demonstrated that recombinant prophage-derived depolymerases with an established enzymatic activity could significantly reduce the mortality of G. mellonella larvae infected with A. baumannii of corresponding K types. In the long term, this means that specific prophage-derived depolymerases, as well as enzymes encoded in lytic phage genomes have great potential as antivirulence agents to control A. baumannii belonging to corresponding capsular types.
The study is published in the International Journal of Molecular Sciences.
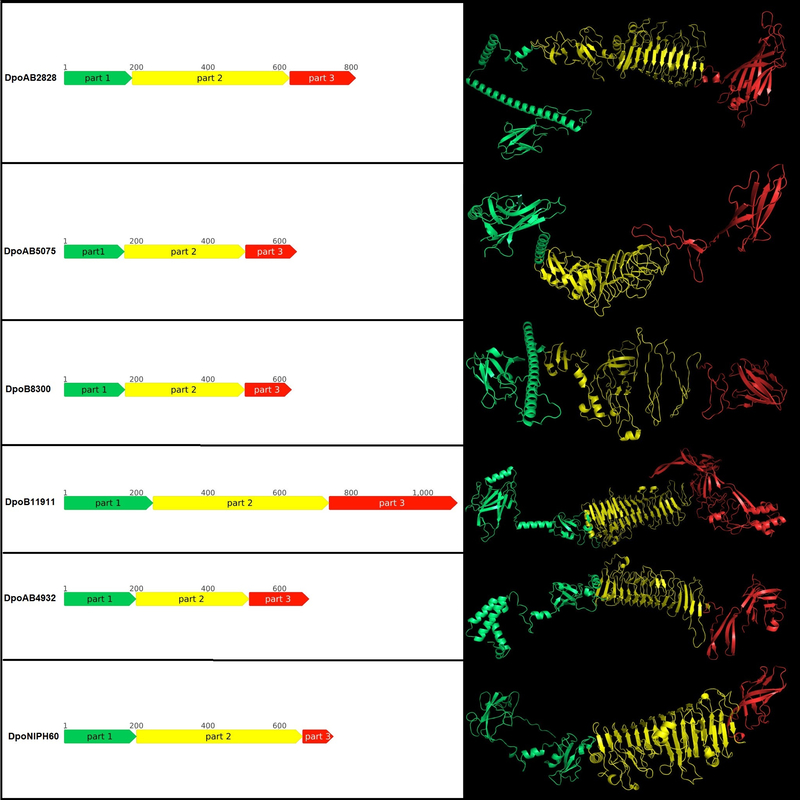
Figure 2. Schematic representation and predicted spatial structure of the prophage-derived depolymerases. The parts of proteins are coloured according to their suggested roles. Part 1 (coloured green) contains the particle-binding N-terminal domain, part 2 (coloured yellow) contains the central pyramidal domain, the part 3 (coloured red) contains C-terminal domain.
june 16, 2022


