Лаборатория протеомики
|
Отдел пептидно-белковых технологий Руководитель: Говорун Вадим Маркович |
Лаборатория протеомики была организована в институте в 2006 году.
Термин «proteome» был предложен в 1995 году Марком Уилкинсом (Marc Wilkins) и соавторами[1] для описания полного набора белков, кодируемых геномом, чуть позже, в 1997 году, Питером Джеймсом был введён в оборот и термин «proteomics»[2]. Сегодня протеомику можно определить как совокупность технологий, направленных на крупномасштабное изучение структур, пост-трансляционных модификаций и взаимодействий белков в живых организмах. В основе этой области исследований лежат базы данных сиквенсов геномов, масс-спектрометрия, как основной инструментальный метод анализа белков и пептидов и биоинформатика, коррелирующая результаты масс-спектрометрического анализа со структурами белков, транслированными из геномных баз данных. Основной задачей протеомики является количественная оценка изменений уровня экспрессии белков в клетках, тканях или целом организме при воздействии на них различных внешних факторов.
В настоящее время в лаборатории развиваются два основных направления исследований:
- Поиск в биологических жидкостях человека пептидно-белковых маркеров социально-значимых заболеваний
- Структурная и функциональная протеомика мха Physcomitrella patens
Поиск в биологических жидкостях человека пептидно-белковых маркеров социально-значимых заболеваний
В качестве одного из наиболее многообещающих аспектов практического приложения результатов исследования протеома человека можно назвать выявление белков и пептидов, содержание которых в организме коррелирует с развитием в организме различных патофизиологических состояний. Поиск новых маркеров для ранней диагностики и идентификация потенциальных молекулярных мишеней для разработки лекарственных препаратов, направленных против социально-значимых заболеваний, являются наиболее очевидными целями подобных исследований. Используя метод сравнительного масс-спектрометрического профилирования образцов сыворотки крови практически здоровых доноров и пациентов с различными заболеваниями (рак яичников, колоректальный рак, доброкачественные гинекологические заболевания, синдром Гийена-Барре, менингиты различной этиологии) нами были построены классификационные модели, различающие масс-спектрометрические профили сывороток крови здоровых людей и пациентов с разнообразными заболеваниями. Специфичность и чувствительность некоторых из таких моделей приведена в Таблице 1.
| Заболевание | Специфичность | Чувствительность |
|---|---|---|
| Рак яичников | 100% | 100% |
| Колоректальный рак | 100% | 100% |
| Сифилис | 92,0% | 100% |
| Аденомиоз | 100% | 93,8% |
В настоящее время фокус биомаркерных исследований нашей лаборатории переместился на детальное изучение пептидомов производных крови и спинномозговой жидкости.
На сегодняшний день, используя разработанный нами метод выделения пептидов, в образцах сыворотки крови практически здоровых людей и пациентов с раком яичников и колоректальным раком нами идентифицировано около 6000 пептидов (Рис. 1).
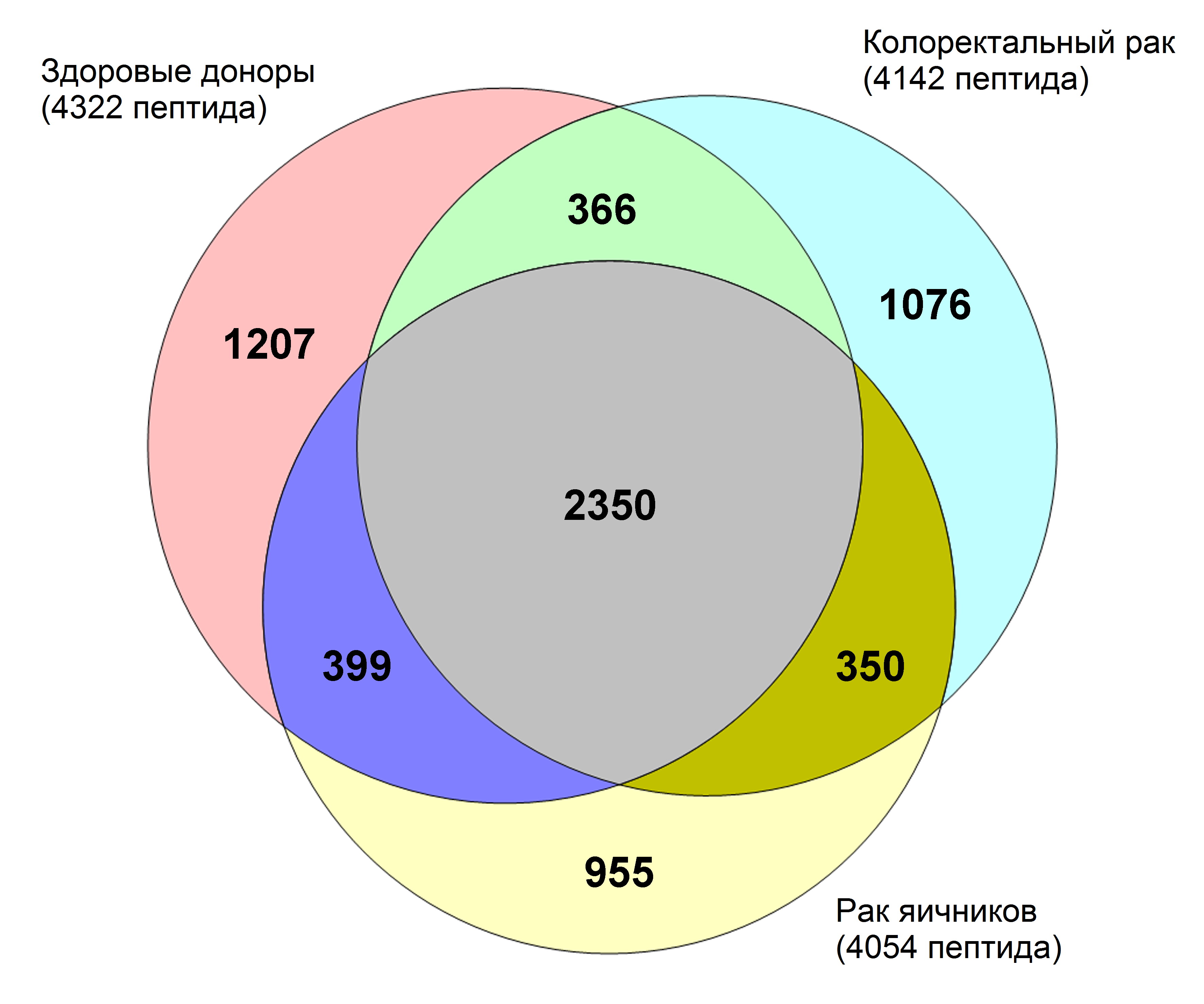
Рисунок 1 – Количество идентифицированных пептидов в образцах сыворотки крови практически здоровых доноров и пациентов с раком яичников и колоректальным раком. Всего было идентифицировано 5840 уникальных пептидов, являющихся фрагментами 877 белков
Из другого объекта наших пептидомных исследований – спинномозговой жидкости, нами выделено и идентифицировано около 2500 пептидов. На Рис. 2 показано сравнение результатов наших исследований пептидома ликвора с результатами аналогичных работ, опубликованных в мировой литературе[3,4].
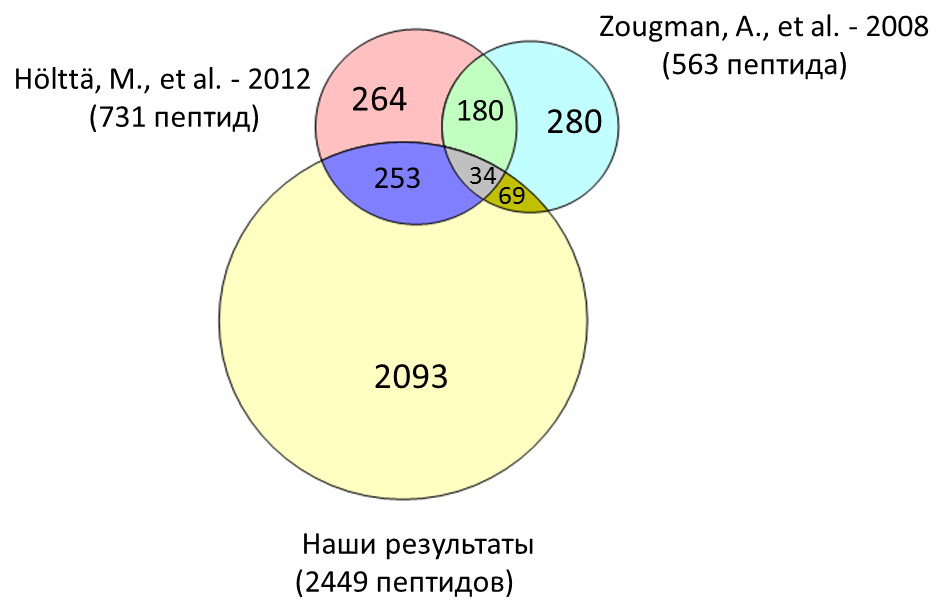
Рисунок 2 – Сравнение результатов работ, проводимых в лаборатории протеомики ИБХ РАН по идентификации пептидов спинномозговой жидкости, с основными результатами аналогичных работ, опубликованных в мировой литературе
Структурная и функциональная пептидомика и протеомика мха Physcomitrella patens
Мхи (Musci, Bryophytaea) являются одними из древнейших представителей флоры Земли. Этим растениям свойственен широкий диапазон биохимической и физиологической пластичности, благодаря которому они способны адаптироваться к экстремальным режимам температуры, влажности и света, а также образовывать ассоциации симбиотического характера с разнообразными организмами, прежде всего, с азотфиксирующими цианобактериями.
Среди представителей этого отдела высших растений, использовавшихся в научных исследованиях, наибольшее распространение в качестве модельного объекта получил вид Physcomitrella patens, выделяющийся среди других мхов целым рядом интересных особенностей. Так, например, Physcomitrella patens является однодомным растением, что позволяет проводить скрещивания in vitro. Кроме того, в отличие от других наземных растений, у этого мха обнаружен высокий уровень гомологичной рекомбинации в ядерном геноме, что даёт возможность проведения генного таргетинга (gene targeting – замещение последовательностей нуклеотидов в составе целевых генов организма путём гомологичной рекомбинации ДНК) с той же эффективностью, что и в дрожжах Saccharomyces cerevisiae.
В 2008 г. усилиями международного консорциума была определена и опубликована полная нуклеотидная последовательность ядерного генома Physcomitrella patens (около 480 млн. нуклеотидных пар оснований), что сделало возможным изучение протеома этого организма. В лаборатории к настоящему времени получены данные по протеому хлоропластов и пептидому различных жизненных форм Physcomitrella patens.
Протеомика хлоропластов Physcomitrella patens
Хлоропласты – органеллы симбиотического происхождения, в которых происходят такие процессы, как фотосинтез, фиксация углерода, биосинтез аминокислот, жирных кислот, крахмала, витаминов, фитогормонов и вторичных метаболитов. При определённых условиях они могут служить источником питательных веществ и играют активную роль в метаболических процессах. Хлоропласты содержат несколько тысяч белков, большинство из которых кодируется ядерным геномом, собственная ДНК хлоропластов кодирует лишь малую часть белков этой органеллы. Продукты ядерных генов синтезируются в цитозоле и, пройдя посттрансляционные модификации, транспортируются в хлоропласты.
Из протонемы Physcomitrella patens нами изолированы интактные хлоропласты и проведён их протеомный анализ. В результате этих исследований было идентифицировано 1445 белков. В силу плохой аннотированности генома Physcomitrella patens большинство идентифицированных белков (96,6%) попали в разряд так называемых предсказанных белков. Их аннотацию проводили путём поиска гомологий с белковыми последовательностями Arabidopsis thaliana с использованием on-line сервиса BLASTP. По субклеточной локализации (по данным Uniprot ) идентифицированные белки распределились следующим образом : хлоропласты - 41,89%, хлоропласты (двойная локализация) – 5,99%, пероксисомы – 1,31%, ядро – 4.49%, митохондрии – 12,46%, мембраны – 1,78%, аппарат Гольджи – 2,15%, глиоксисомы - 0,18%, эндоплазматический ретикулум - 3,75%, цитоплазма – 15,97%, клеточная мембрана – 5,53%, секреторные – 2,72%, вакуоль – 1,31%, неизвестная локализация 0,47%.
Дальнейшая работа по изучению протеома хлоропластов будет направлена на исследование пост-трансляционных модификаций хлоропластных белков, а также на их количественные изменения в ответ на стрессовые воздействия.
Изучение пептидома клеток мха Physcomitrella patens
Роль пептидов в регуляции процессов роста и развития, в защитных реакциях и сигнальных процессах высших растений в последние годы привлекает все большее внимание исследователей (Matsubayashi and Sakagami, 2006; Farrokhi et al., 2008; Murphy et al., 2012). Известные на сегодняшний день биоактивные пептиды растений регулируют такие ключевые процессы, как активность апикальных побеговых и корневых меристем, закладка элементов сосудистой системы при делении клеток камбия и формирование устьичного аппарата в процессе дифференциации клеток листового эпидермиса, симбиоз и пыльцевую самонесовместимость (Farrokhi et al., 2008) (Katsir et al,. 2012). Чаще всего биоактивные пептиды образуются в результате специфического протеолиза белков-предшественников. Однако, в клетках и тканях растений могут содержаться пептидные пулы, образующиеся в результате процессов деградации клеточных белков, которые также могут обладать активностью [5]. Кроме того, в последнее время в ряде исследований показано наличие в клетках биоактивных пептидов, транслирующихся с коротких рамок считывания (sORF) [6]. Знания о растительных пептидах важны не только для системной биологии, но могут использоваться в различных прикладных областях, в сельском хозяйстве, в производстве диетических продуктов и фармацевтических препаратов. В настоящее время многие публикации по пептидомике растений представляют работы, нацеленные на поиск и идентификацию пептидов с определённой, например, антимикробной, биологической активностью.
Целью нашей работы является изучение пептидома модельного объекта растительной биологии – мха Physcomitrella patens.
Мох Physcomitrella patens - новый модельный объект биологии растений [7,8,9]. Секвенирование ядерного генома P. patens [10]значительно повысило эффективность системных подходов при изучении физиологических и биохимических процессов. В последние годы активно изучались протеом [11,12,13,14,15,16,17], транскриптом [18,19,20,21,22]и метаболом [23]клеток мха как в стандартных условиях роста, так и при различных стрессовых воздействиях. Однако работ в области пептидомики проводится мало. Это связано со сложностями, при обнаружении и анализе новых эндогенных пептидов растений, это зачастую связано с рядом трудноразрешимых методических проблем. Они связаны с небольшим содержанием пептидов в клетках и тканях растений на фоне значительного количества балластных растительных соединений, например, фенолов, с которыми пептиды весьма трудно разделимы физико-химическими методами. Мы использовали гаметофоры, протонему и протопласты мха для выделения и анализа пула эндогенных пептидов, являющихся продуктом деградации функционально активных белков клетки. А также для системного анализа процессов пептидогенеза в растительной клетке.
В результате проведения масс-спектрометрического анализа пептидных образцов, в гаметофорах мха идентифицировано около 4000 тысяч эндогенных пептидов, являющихся фрагментами 761 белка. Были идентифицированы фрагменты ряда хлоропластных белков, а также некоторых хорошо известных стрессовых белков, например late embryogenesis abundant protein (LEA). Помимо LEA белков, выявлено большое количество уникальных эндогенных пептидов, фрагментов таких основных белков как RUBISCO, пластоцианин, фактор элонгации 1 альфа и ряд других. В клетках протонемы мха идентифицировано около 4000 уникальных пептидов, являющихся фрагментами 855 белков-прекурсоров. Белки-прекурсоры, представленные большим количеством пептидов, относились к основным белкам клетки, например большая субъединица RUBISCO, фактор элонгации 1-альфа и ряд других. При сравнительном анализе пептидогенных белков гаметофоров и протонемы мы идентифицировали 270 уникальных для протонемы белков-прекурсоров и около 3300 пептидов. Следует отметить большую схожесть гаметофоров и протонемы по белкам-прекурсором, чем по пулу пептидов (Рисунок 3).
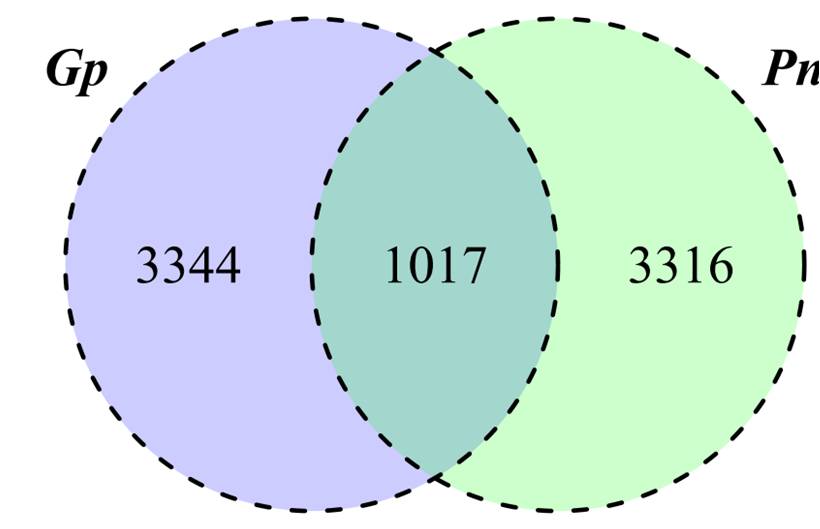
Рисунок 3 – Сравнительный анализ содержания количества нативных пептидов в протонеме (Pn) и гаметофитафорах (Gp)
В качестве объекта для анализа влияния стрессовых условий на пептидом клетки были выбраны протопласты мха P. patens. Для выделения протопластов из протонемы был использован ферментный препарат драйзелаза (Driselase), получаемый из базидиомицетов. Для контроля потенциальной протеолитической активности драйзелазы, в растворе для выделения протопластов проведено инкубирование БСА (Бычий сывороточный альбумин). Установлено, что драйзелаза не обладает самостоятельной протеолитической активностью, которая может приводить к артефактам при выделении. В пептидоме протопластов, было обнаружено 20 427 уникальных пептида, являющихся фрагментами 1572 белков. Количество уникальных пептидов в протопластах при этом возрастает в несколько раз по сравнению с протонемой и гаметофорами. При этом, около 19000 уникальных пептидов идентифицировались только в протопластах (Рисунок 4).
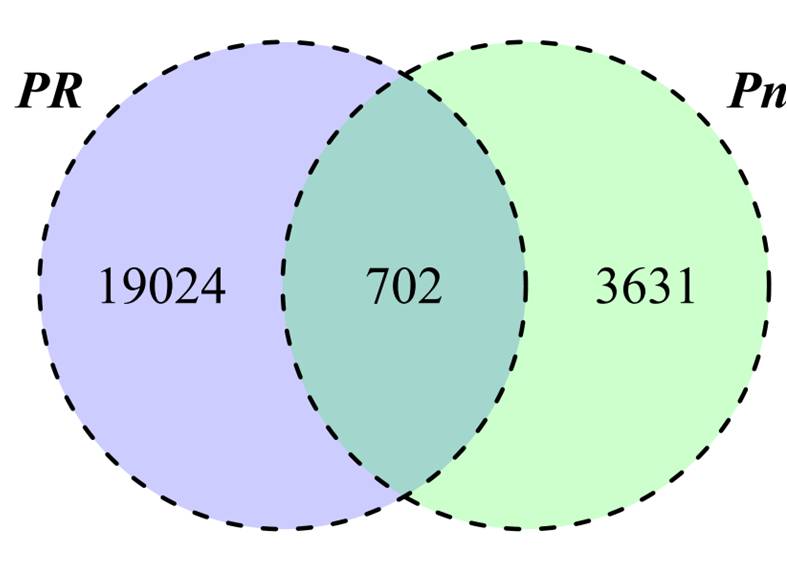
Рисунок 4 - Сравнительный анализ содержания количества эндогенных пептидов в протонеме (Pn) и протопластах (Pr)
Анализ транскрипции некоторых белков, выявленных в протопластах, указывает на то, что в сравнении с протонемой экспрессия соответствующих генов практически не меняется. Эти данные, по-видимому, указывают на то, что повышенная деградация этих белков не связана с увеличением их представленности в клетке. Кроме того, было идентифицировано 1179 пептидогенных белка, фрагменты которых обнаруживаются только в протопластах, среди них 263 белка были представлены 10 и более пептидами.

Лаборатория протеомики. В нижнем ряду (слева направо): асп. Аниканов Н.А., к.б.н., м.н.с. Середина А.В., д.б.н., профессор, зав. лаб. Говорун В.М., асп. Иванова О.М., к.х.н., с.н.с. Зиганшин Р.Х., асп. Хазигалеева Р.А., асп. Шендер В.О. В дальнем ряду: к.х.н., н.с. Ковальчук С.И., сотр. Азаркин И.В., сотр. Арапиди Г.П., к.б.н., н.с. Фесенко И.А.
Литература
1. Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, et al. (1996) Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev 13: 19-50.
2. James P (1997) Protein identification in the post-genome era: the rapid rise of proteomics. Q Rev Biophys 30: 279-331.
3. Holtta M, Zetterberg H, Mirgorodskaya E, Mattsson N, Blennow K, et al. (2012) Peptidome analysis of cerebrospinal fluid by LC-MALDI MS. PLoS One 7: e42555.
4. Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, et al. (2008) Integrated analysis of the cerebrospinal fluid peptidome and proteome. J Proteome Res 7: 386-399.
5. Gara OG, Iatskin ON, Shvets VI, Karelin AA, Ivanov VT (2006) [Isolation and structure of peptides from oat (Avena sativa) seedlings]. Bioorg Khim 32: 211-220.
6. Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, et al. (2013) Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 9: 59-64.
7. Kamisugi Y, Schlink K, Rensing SA, Schween G, von Stackelberg M, et al. (2006) The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res 34: 6205-6214.
8. Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci U S A 95: 4368-4373.
9. Decker EL, Frank W, Sarnighausen E, Reski R (2006) Moss systems biology en route: phytohormones in Physcomitrella development. Plant Biol (Stuttg) 8: 397-405.
10. Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64-69.
11. Sarnighausen E, Wurtz V, Heintz D, Van Dorsselaer A, Reski R (2004) Mapping of the Physcomitrella patens proteome. Phytochemistry 65: 1589-1607.
12. Cho SH, Hoang QT, Kim YY, Shin HY, Ok SH, et al. (2006) Proteome analysis of gametophores identified a metallothionein involved in various abiotic stress responses in Physcomitrella patens. Plant Cell Rep 25: 475-488.
13. Skripnikov AY, Polyakov NB, Tolcheva EV, Velikodvorskaya VV, Dolgov SV, et al. (2009) Proteome analysis of the moss Physcomitrella patens (Hedw.) B.S.G. Biochemistry (Mosc) 74: 480-490.
14. Wang X, Yang P, Gao Q, Liu X, Kuang T, et al. (2008) Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 228: 167-177.
15. Wang X, Yang P, Zhang X, Xu Y, Kuang T, et al. (2009) Proteomic analysis of the cold stress response in the moss, Physcomitrella patens. Proteomics 9: 4529-4538.
16. Cui S, Hu J, Guo S, Wang J, Cheng Y, et al. (2012) Proteome analysis of Physcomitrella patens exposed to progressive dehydration and rehydration. J Exp Bot 63: 711-726.
17. Lang EG, Mueller SJ, Hoernstein SN, Porankiewicz-Asplund J, Vervliet-Scheebaum M, et al. (2011) Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep 30: 205-215.
18. Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS (2007) Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol 176: 275-287.
19. Richardt S, Timmerhaus G, Lang D, Qudeimat E, Correa LGG, et al. (2010) Microarray analysis of the moss Physcomitrella patens reveals evolutionarily conserved transcriptional regulation of salt stress and abscisic acid signalling. Plant Molecular Biology 72: 27-45.
20. Xiao L, Wang H, Wan P, Kuang T, He Y (2011) Genome-wide transcriptome analysis of gametophyte development in Physcomitrella patens. BMC Plant Biol 11: 177.
21. Xiao L, Zhang L, Yang G, Zhu H, He Y (2012) Transcriptome of protoplasts reprogrammed into stem cells in Physcomitrella patens. PLoS One 7: e35961.
22. Nishiyama T, Miyawaki K, Ohshima M, Thompson K, Nagashima A, et al. (2012) Digital gene expression profiling by 5'-end sequencing of cDNAs during reprogramming in the moss Physcomitrella patens. PLoS One 7: e36471.
23. Erxleben A, Gessler A, Vervliet-Scheebaum M, Reski R (2012) Metabolite profiling of the moss Physcomitrella patens reveals evolutionary conservation of osmoprotective substances. Plant Cell Rep 31: 427-436.
| ФИО | Должность | Контакты |
|---|---|---|
Ранее здесь работали | ||
| Грачёв С.А., к.х.н. | ||
| Кадыков В.А., д.б.н. | ||
| Клинов Д.В., к.ф.-м.н. | ||
| Середина А.В., к.б.н. | ||
| Азаркина (Хазигалеева) Регина Айдаровна | ||
| Аниканов Николай Андреевич | ||
| Гаранина И.А. | ||
| Иванова Ольга Максимовна | ||
| Пушкова Е.Н. | ||
| Дёмин В.В., к.х.н. | ||
| Логвина Н.А., к.х.н. | ||
 Загрузка...
Загрузка...Научные проекты
 Загрузка...
Загрузка...Говорун Вадим Маркович
Москва, ул. Миклухо-Маклая, 16/10 — На карте
 Загрузка...
Загрузка...
