Press-room / news / Science news /
Identification of animal venoms by Raman spectroscopy combined with principal component analysis
Scientists from the Department of Molecular Neuroimmune Signaling of the Institute of Bioorganic Chemistry RAS, together with colleagues from the Prokhorov General Physics Institute RAS using Raman spectroscopy carried out a comparative analysis of various animal venoms and a number of different nature substances (not-venoms), and the applicability of this method for the identification of animal venoms was demonstrated.
The venoms of various animals, including snakes, scorpions, and spiders, are complex mixtures of predominantly protein and peptide toxins and have a wide range of effects, often adverse, on living organisms. However, venoms and their components are used, for example, in the pharmaceutical and cosmeceutical industries. Due to the limited sources and difficulty of obtaining, some venoms are highly valuable. The smuggling of wildlife products, including animal venoms, is a global problem today, and their illegal trafficking is significant. It is often impossible to prove that the transported material is an animal venom, that allows smugglers to avoid responsibility.
Raman spectroscopy, when applied to protein substances, offers advantages such as high information yield and the ability to directly measure dry samples (without preparation) in small quantities. This method can be used to implement a quick and simple methodology for identifying animal venoms.
We demonstrated that Raman spectroscopy combined with principal component analysis (PCA) can distinguish between dry samples of crude venoms and not-venoms considered; the clustering of different animal venom samples based on their similarities was observed as well. Snake, scorpion, spider, bee, and centipede venoms were analyzed. Not-venoms included amino acids, protein and peptide toxins (HDP1, NT1, Az), and the proteins pepsin and lysozyme. The resulting PCA map, in which the distance between samples reflects their differences, is shown in Figure. Interestingly, the relative similarity between scorpion and spider venoms is visible in the map.
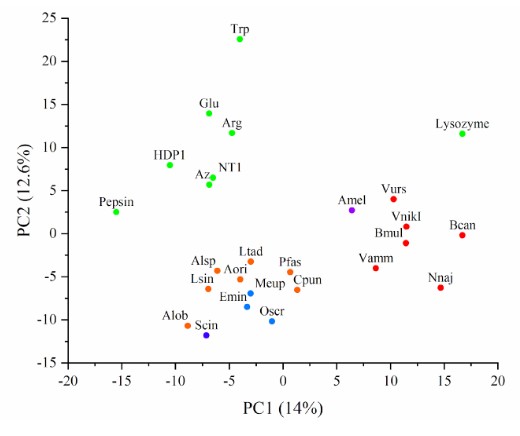
Figure. PCA score plot for the analyzed samples. Percentage of explained variance for each principal component (PC) is indicated in brackets. Samples of different nature differ in colour. Not-venom samples are in green; for venoms: orange – spiders, light blue – scorpions, blue – scolopendra, red – snakes, purple – bee.
Machine learning algorithms, including training on a subset of samples, also enabled the automatic classification of test substances into venoms and not-venoms.
The proposed methodology appears promising for on-site identification of animal venoms, for example, at airports and border control by staff without special education. Creating libraries and software on a PC coupled with a compact Raman spectrometer can serve as the basis for semi-automated identification of suspected substances.
It should be noted that further studies with an expanded sample set, including non-protein organic substances, carbohydrates, salts, etc., are necessary to confirm the effectiveness of the proposed method. This work was carried out in collaboration between researchers from biological and physical teams, aimed at advancing optical methods, one of which is Raman spectroscopy, in the field of biology, in particular, toxinology. This work was performed within the framework of the State assignment, theme number 124051400034-0.
november 11, 2025


