Laboratory of mechanisms of gene expression
The Laboratory of Mechanisms of Gene Expression was organized in May of 2007 on the basis of an independent research group under the same name existed since December 2001.
Since the very beginning the main focus of our research is studies of basic mechanisms of transcription in eukaryotes. The functional complementations in vivo of certain subunits of nuclear RNA polymerases I—III among evolutionary remote species (yeast, drosophila, man) has been first demonstrated (Gene, 1994, 147: 63–69; Mol. Cell. Biol., 1995, 15: 4702–4710) and functional relatedness of small subunits of RNA polymerases in Archaea and Eucarya was established (Russ. J. Bioorg. Chem., 1997, 23: 100–106;J. Biol. Chem., 1999, 274: 8421–8427).
The first comprehensive studies of exchangeability in vivo (in the cells of yeast Saccharomyces cerevisiae) of the subunits of nuclear RNA polymerases I, II, and III between evolutionary distant species (Schizosaccharomyces pombe and Homo sapiens) have been performed, whereby many of the components of the basal transcription apparatus of the fission yeasts were cloned and thoroughly characterized (J. Mol. Biol., 2000, 295: 1119—1127; Mol. Biol. (Mosk), 2002, 36: 3—26;Nucleic Acids Res., 2006, 34: 3615—3624). As a part of the the International Project on total sequencing of the Sch. pombe genome in three loci of its chromosome I the primary structure gaps were closed and the nucleotide sequence continuity was restored with clones pYUK71, pYUL23 and pYUG7 isolated in our laboratory (Nature, 2002, 415: 871—880; see also S. pombe genetic map). In the collaborative work with French scientists from ESBS (Strasbourg) was found that one of the subunits of human RNA polymerase II is encoded by a multigene family which expression give rise to at least three protein isoforms (BMC Mol. Biol., 2001, 2:14). Four independent genes encoding various variants of this hRPB11 subunit were revealed (Russ. J. Bioorg. Chem., 2004, 30: 561—565). Coevolution of Homo sapiens POLR2J and PMS2 genes, both located on chromosome 7 and encoding two essential components of such important molecular processes of the living cell as transcription and DNA mismatch repair, was shown: both these gene families undergone multiple rounds of duplication events started in the hominoid ancestor less than 20 million years ago (Dokl. Biochem. Biophys., 2006, 408: 175—179). Moreover, according to data obtained the evolution of these two gene families includes stages specific for humans (Russ. J. Genet., 2010, 46: 1112-1114; Biochemistry, 2011, 76: 976-980; Cell & Tissue Biology, 2013, 7: 314-319).
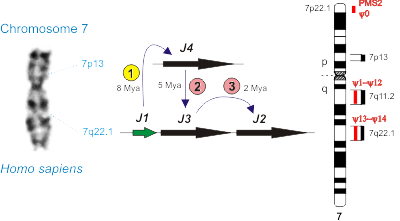
Mapping on the human chromosome 7 several genes that are important for primate evolution: four POLR2J paralogues (J1–J4) and sixteen PMS2 paralogues (PMS2, ψ0, ψ1–ψ14). Different stages (1, 2 and 3) of POLR2J amplification are shown; two of them are specific for Homo sapiens (marked with pink circles). Mya — million years ago.
| Fullname | Position | Contacts |
|---|---|---|
Previously worked here | ||
| Klykov V.N., Ph.D. | ||
| Proshkin S.A. | ||
| Sharshavina E.A. | ||
| Shpakovski D.G. | ||
| Slovokhotov I.Y. | ||
 Loading...
Loading...Scientific projects
 Loading...
Loading...Shpakovski G.V.
Russia, Moscow, Ul. Miklukho-Maklaya 16/10 — On the map
 Loading...
Loading...
