Press-room / news / Science news /
«Transcriptomics mycobacteria as a way to study the mechanisms of control of latent tuberculosis»
Until now, scientists do not fully understand what is happening in the cells of M. tuberculosis in a resting stage. However, the answer to this question is an absolute must, because without it is impossible to find ways to defeat tuberculosis. Employees of Laboratory of human genes structure and functions M.M. Shemyakin–Yu.A. Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences T. L. Azhikina and D.V. Ignatov studied the transcript of the genus Mycobacterium and identify possible molecular mechanisms involved in establishing and maintaining a state of rest of the bacterial cells. The results of this work presented D.V. Ignatov on X International Workshop Conference for Young Researchers «Current Aspects of Modern Microbiology».
Since the bacterium Mycobacterium tuberculosis is already familiar to mankind some 5,000 years, but so far medicine cannot completely defeat tuberculosis - a disease that causes the organism. The fact tuberculosis infection begins with getting M. tuberculosis in human lungs. At the same time quickly formed, an immune response and a significant portion of the bacteria are absorbed by pulmonary macrophages. However, since the microorganisms have the ability to survive inside the macrophage, some bacteria remain unharmed. Thus, it does not reach to the complete destruction of pathogens, and it goes into a latent form.
Macrophages, in turn, can inhibit the growth of bacteria, making their environment more acidic, and that M. tuberculosis rather dislikes. Thereafter, infected macrophages migrate into the inner tissues of the lung where they develop around the granuloma - a set of different immune cells. These granulomas can exist for several years - this corresponds to the stage of latent tuberculosis infection. Themselves bacteria inside the granuloma turn into dormant state during which they almost fall, and the level of their metabolism is low. However, while in this state, they are little susceptible to many external factors - including drugs.
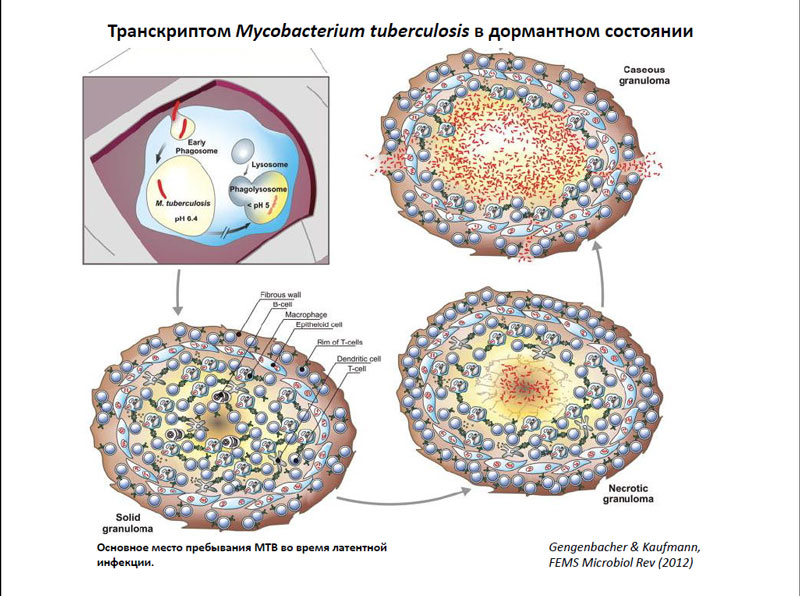
Transcriptome Mycobacterium tuberculosis in the dominant state
To answer the question, what exactly happens in the cells of M. tuberculosis within the granuloma, it is difficult, because there is no animal model, which can accurately reproduce the granuloma, and the number of bacterial cells within it is very small. However, the answer to this question is an absolute must, because without it, the scientists never be able to find ways to defeat tuberculosis. However, now we developed a variety of an in vitro model, using which you can transfer the bacteria dormant state in order to learn what is happening at this time within the cells of M. tuberculosis.
That such a study was conducted at the Laboratory structure and function of human genes IBCh RAS T. L. Azhikina and D. V. Ignatov, who studied the genus Mycobacterium transcriptomics, together with research teams from the Federal Research Center «Fundamentals of Biotechnology» RAS and the Central Scientific Research Institute of Tuberculosis RAMS (both - Moscow). In the process, it has been clarified many interesting points related to the transcriptome of mycobacteria.
What is a transcript? This includes all of the RNA present in the bacterial cell. Interestingly, more than 80 percent of the ribosomal RNA transcriptome up. Share the same mRNA, although it may vary depending on growth phase is generally small - less than 5 percent. Everything else - is transport (10 percent), and various regulatory RNA. The main types of regulatory RNAs in bacteria are riboswitches and siRNAs. Riboswitches arranged mRNA in untranslated regions of genes. They work as follows - when bound with them any low molecular weight substances begins activation or inhibition of translation of these same genes. Unlike riboswitches, siRNAs are transcribed independently by their regulatory genes. Small interfering RNAs complementary interaction with mRNA leads either to activation or to the inhibition of translation of these genes.
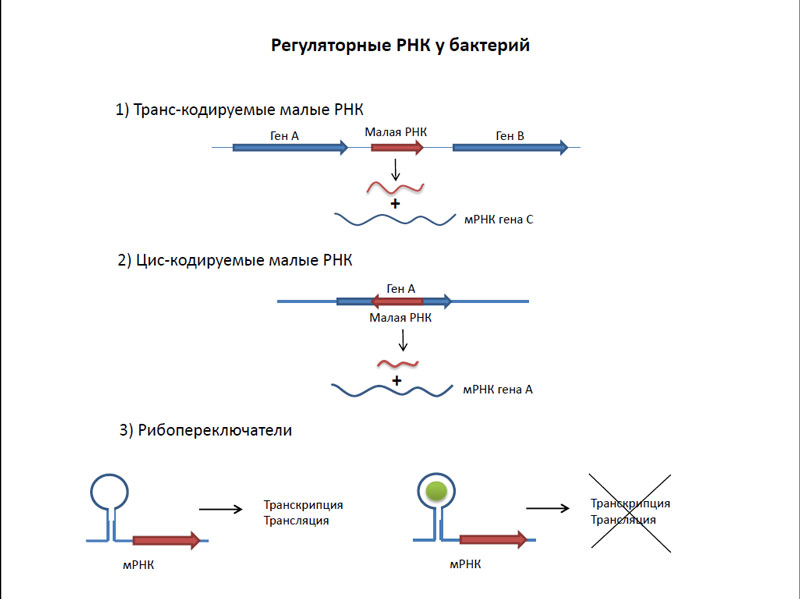
Bacteria regulatory RNA
Thus, studying the transcript can identify which genes are active in a cell at the moment, and which, on the contrary, do not work. In addition, these studies give an idea of how to regulate many processes in the cells of microorganisms. This, in turn, will help you understand exactly how the Mycobacterium cell becomes dormant state and what happens to it at this stage of the life cycle.
In the study, researchers used transcriptome mycobacteria RNA-seq, based on the sequencing of the cDNA fragments using the next generation sequencers. Using the method of RNA-seq can search for new genes in bacteria, genes including regulatory and non-coding RNAs with extreme accuracy (up to one nucleotide) mapping boundary transcripts measuring transcription levels of genes in a wide range of structures and determine operons.
The essence of this method consists in the following - is first bacterial cells are isolated RNA. Then it was necessary to get rid of the most numerous, but it is not interesting to our study of the ribosomal RNA. The residue is then fragmented and attached to each fragment of the 5 'and 3' terminal adapters, which are needed for the seed cDNA synthesis. After this the synthesis and amplification of cDNA, and then - these DNA sequencing using the next generation sequencers. Based on the resulting sequencing data card constructed genes of bacteria that were analyzed using bioinformatics methods.
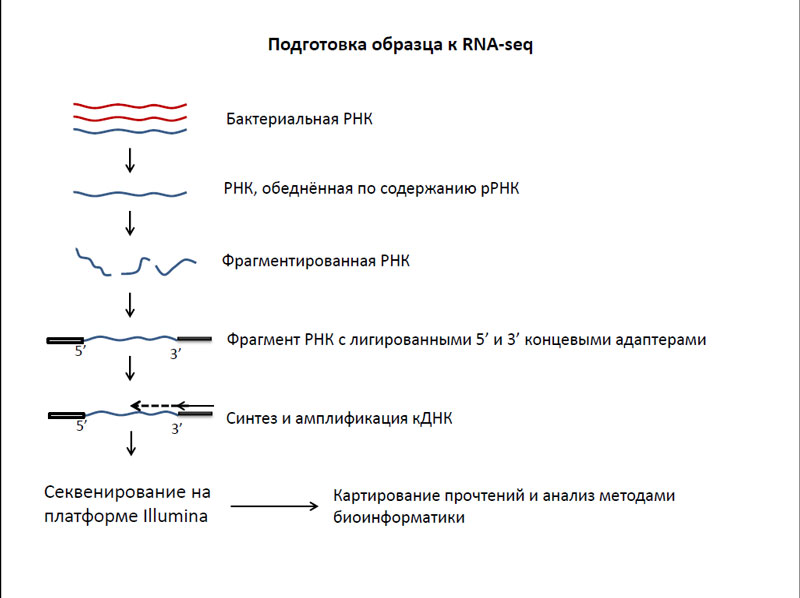
RNA-seq
The objects of the study were in the cell Mycobacterium avium and Mycobacterium tuberculosis - the last at the same time were in a dormant state. According to D. Ignatov, «when we started our work, then there is only one such study is conducted on cells of M. tuberculosis in culture. Now there are about a dozen studies on the transcriptome of these bacteria. It says that so far the processes occurring in the cells of mycobacteria in a resting stage, studied bad enough».
Infection with the pathogen Mycobacterium avium gives rise to a number of diseases in humans and animals. For subspecies M. avium avium and M. avium silvaticum cause disease of birds, and a subspecies of M. avium paratuberculosis - Jones disease of ruminants. For a man the greatest danger is a subspecies of M. avium hominissuis, which is widely distributed in the environment, which if it enters the human body can cause disseminated lung disease in people with immune deficiency. Pathogenesis of Mycobacterium avium is similar to that of M. tuberculosis - the microorganism is also capable of focusing inside the macrophages. Therefore, the study of the transcriptome of the organism can help to understand what processes occur in dormant cells of M. tuberculosis, which are difficult to study in vivo.
After sequencing, the transcriptome Mycobacterium avium was obtained tens of millions of readings, some of which turned out to be ribosomal RNA loci - and this despite the fact that it sought to remove. Part loci mapped in the mRNA part - in intergenic regions (and such was even more than broadcast loci), and part of them was antisense mRNA sequences parts. Obviously, the latter two are related to reading siRNA. As a result, the researchers were able to identify several siRNA transcriptome of M. avium and compare them with those of M. tuberculosis. It turned out that most of these siRNAs have homologues in M. tuberculosis except for a few of them. Thus, in the genome of M. avium lack of homology of two siRNA M. tuberculosis, such as MTS0479 (expectation operates when the cell is experiencing oxidative stress) and MTS1338 (synthesized during stationary phase and hypoxia). In turn, no homologues of M. tuberculosis four siRNA M. avium, whose function is not known.
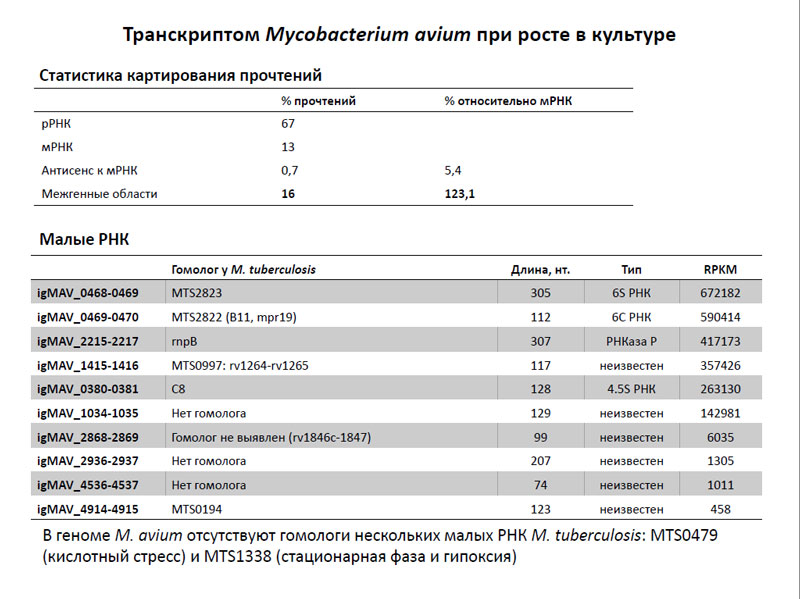
Transcriptome Mycobacterium avium when grown in culture
The second source of reads loci were 5'-untranslated regions. The researchers developed an algorithm for mapping of transcription start sites (TSS) at the transcriptional profile. Moreover, it turned out an interesting thing - 33% TSS coincide with the start codon, that is, they match leaderless mRNA. This is unusual for bacteria, which are mRNAs are very rare - they are known for only a few dozen genes. It is not clear why there were so many in mycobacteria - according to the authors of this issue requires further detailed study. Several also sufficiently long 5'-untranslated regions correspond riboswitches different genes.
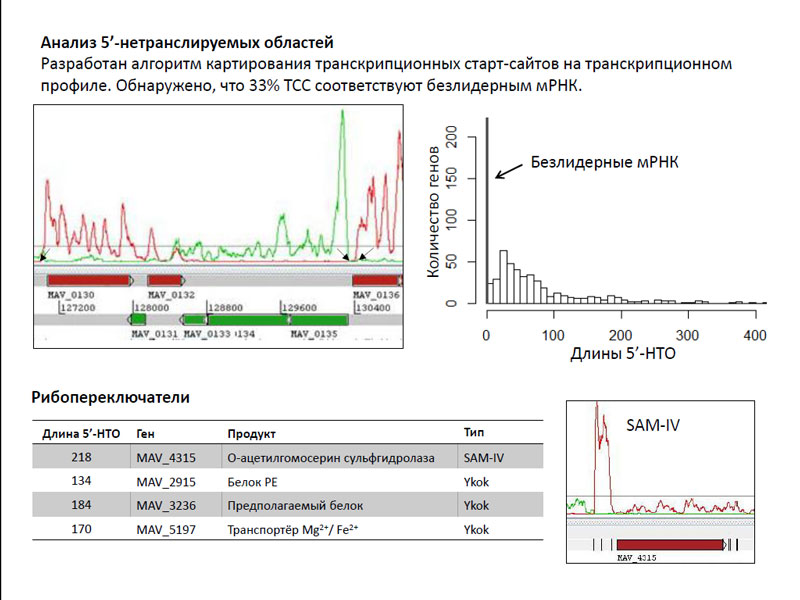
Analysis of the 5'-untranslated regions
Mycobacterium tuberculosis in a dormant state were conducted on a model that was developed in the laboratory of prof. A. S. Kaprelyantz of the A. N. Bach Institute Biochemistry of RAS. M. tuberculosis cells were cultured in a liquid medium, wherein no potassium ions. The grown bacteria thus acquired very interesting properties: they have lost the ability to grow on solid medium, and were insensitive to the effects of several types of antibiotics. Furthermore, these microorganisms have unusual morphology of cells, they have been lowered ATP synthesis and respiration, and was reduced lipid content. As for the level of transcription, it was also low. After two days of such cultivation in the colony of M. tuberculosis were added antibiotic rifampicin inhibitory concentration (5 μg / ml), which kills all actively dividing cells. Thus, only in the culture remained dormant microorganisms.
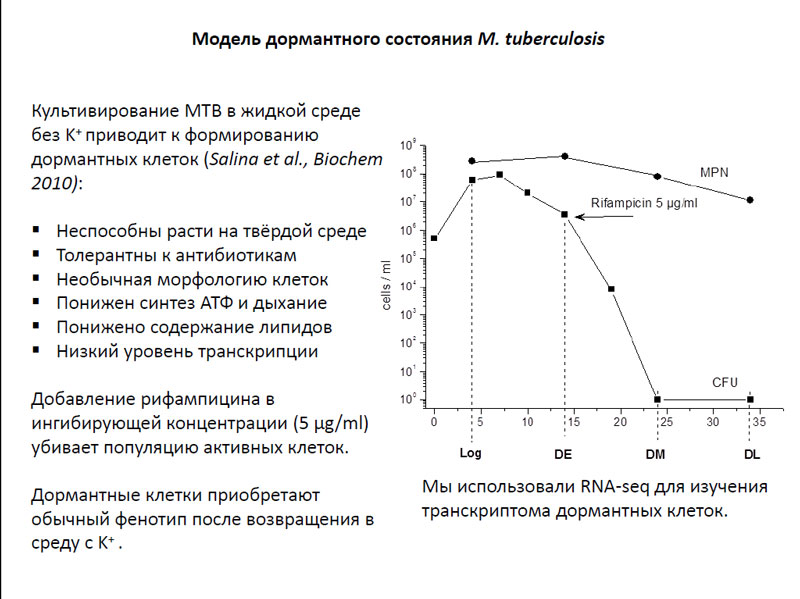
Model dominant state M. tuberculosis
T. L. Azhikina and D. V. Ignatov studied transcriptomics M. tuberculosis cells in the early dormant stage, before the addition of rifampicin, as well as those that were in the late stages of dormant state, which came a few days after antibiotic injection. An interesting feature of the transcriptome dormant cells was cut 23S ribosomal RNA. The researchers were able to map the point of cutting the 23S rRNA - it is located far enough away from the catalytic center of the ribosome, and, apparently, has no effect on protein synthesis. However, about what causes this cutting agent and whether cutting of the physiology of the bacterial cell is not known. However, it is possible that this mechanism is actively promoting the transition of bacterial cells in the dormant state, and supports it.
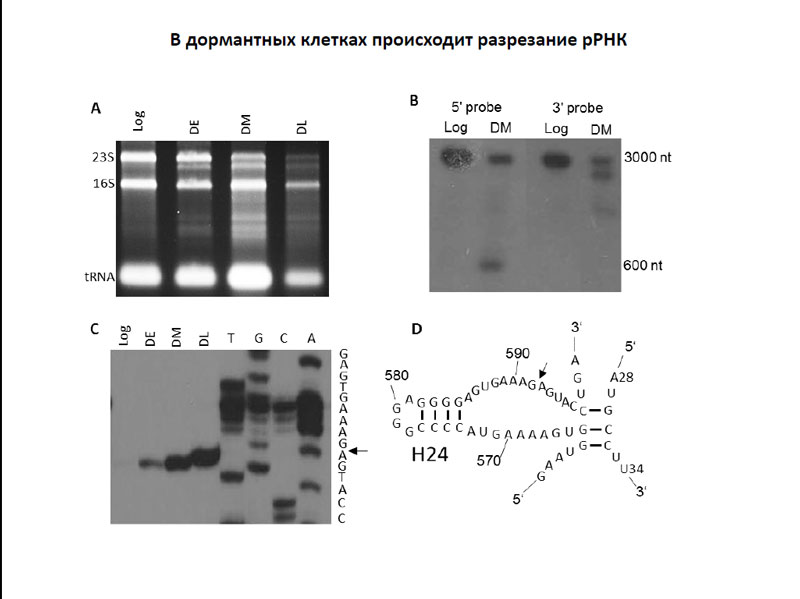
Cutting rRNA in dormant cells
Furthermore, it was found that the dormant cells significantly reduced the level of transcription (this was particularly noticeable in the dormant cells at late stage), and the total amount of mRNA is reduced by 30-50 times as compared with the early stage of dormant. However, despite such a small amount of mRNA, the researchers were able to measure the level of expression of protein-coding genes. The authors focused their attention on those genes that change their expression when cultured in medium without the potassium ions, as well as those whose expression varies by the addition of rifampicin.
The results showed that when cultured in a medium without potassium ions significantly decreased the expression of genes whose proteins are involved in breathing, as well as gene subunit of ATP-synthase. Among the genes whose activity was increased in the absence of potassium ions, it should be noted genes PE-PGRS, which are many in mycobacteria, although it is still their function is unclear. It is only known that these genes in the genome of Mycobacterium spread relatively recently, and first they are specific to pathogenic mycobacteria.
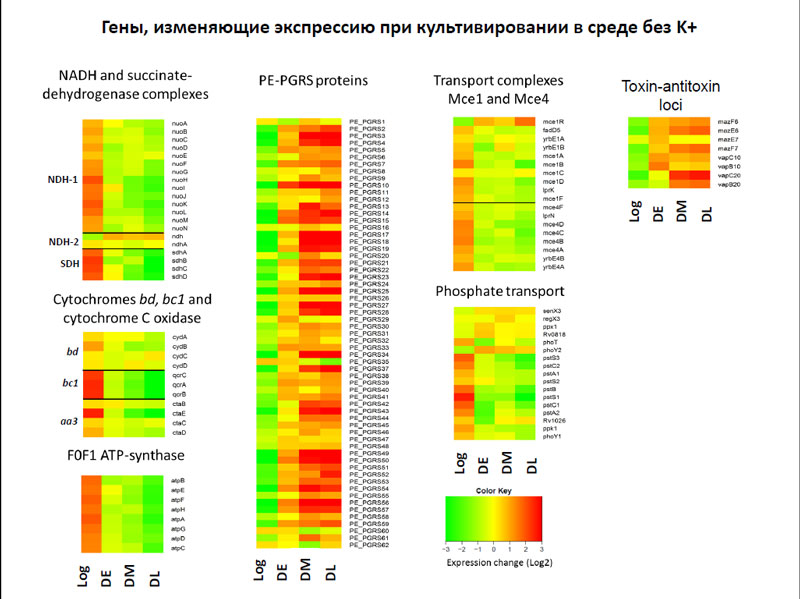
Genes, that alter expression when cultured in medium without K +
After the addition of rifampicin in the cells of the bacteria, reduce the activity of genes encoding enzymes of the tricarboxylic acid cycle and ribosomal proteins.
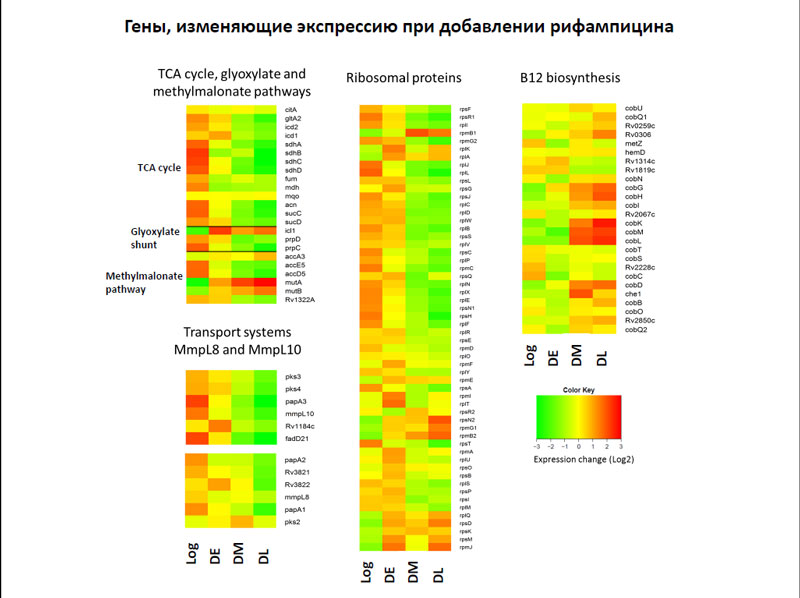
The genes, that change expression with the addition of rifampicin
It was also examined the expression of several siRNA in cells in the early and late dormant state. It was noted that the level of expression of siRNAs, MTS0997, MTS2823 MTS1338 and even dormant bacteria remains high. Interestingly, the expression is controlled MTS1338 DosR transcription factor, which regulates genes involved in the transition to the dormant state. Based on these data, the researchers suggested that it is this siRNA may play a big role in dormant state.
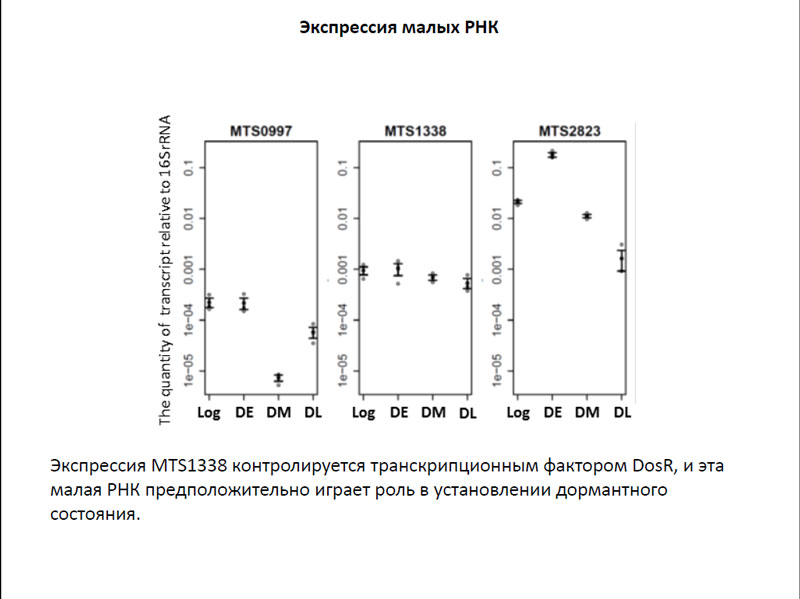
The expression of siRNAs
Thus, T. L. Azhikinа and D. V. Ignatov were first obtained data about the changes taking place in the transcriptome Mycobacterium tuberculosis in the transition to dormant state of the in vitro conditions of potassium deficiency. It was also suggested possible mechanisms involved in the establishment and maintenance of a state of rest. All this can help develop ways to combat the causative agent of tuberculosis in the stationary phase.
References
1) Ignatov, D., Malakho, S., Majorov, K., Skvortsov, T., Apt, A. and Azhikina, T. (2013) RNA-Seq analysis of Mycobacterium avium non-coding transcriptome. PloS one, 8, e74209.
2) Ignatov D., Salina E., Fursov M., Skvortsov T., Azhikina T., Kaprelyants A. (2015). “Dormant non-culturable Mycobacterium tuberculosis retains stable low-abundant mRNA”. BMC Genomics, in press.
november 6, 2015

