Press-room / news / Science news /
Researchers from IBCh found out how the protein isoforms diverse
Researchers of the IBCh RAS and the Federal Research and Clinical center of Physical-Chemical Medicine of FMBA of Russia found that alternative splicing of messenger RNAs makes a non-significant contribution to the plant cell proteome diversity. The results have been published in the Scientific Reports journal.
"When we began the study, we aimed to explore the functions of alternative forms of protein translated from mRNA isoforms that are formed as a result of alternative splicing. However, during the study of cell proteome we did not find as many protein isoforms as it is predicted from alternative splicing. Apparently, the process of alternative splicing is not so important for the production of new proteins, as it is written in textbooks. From another side alternative splicing plays essential role in many other molecular processes. For example we demonstrated that alternative splicing significantly changes mRNA-lncRNA interaction patterns.” says Igor Fesenko, Ph.D., researcher of the Laboratory of Proteomics of the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry RAS.
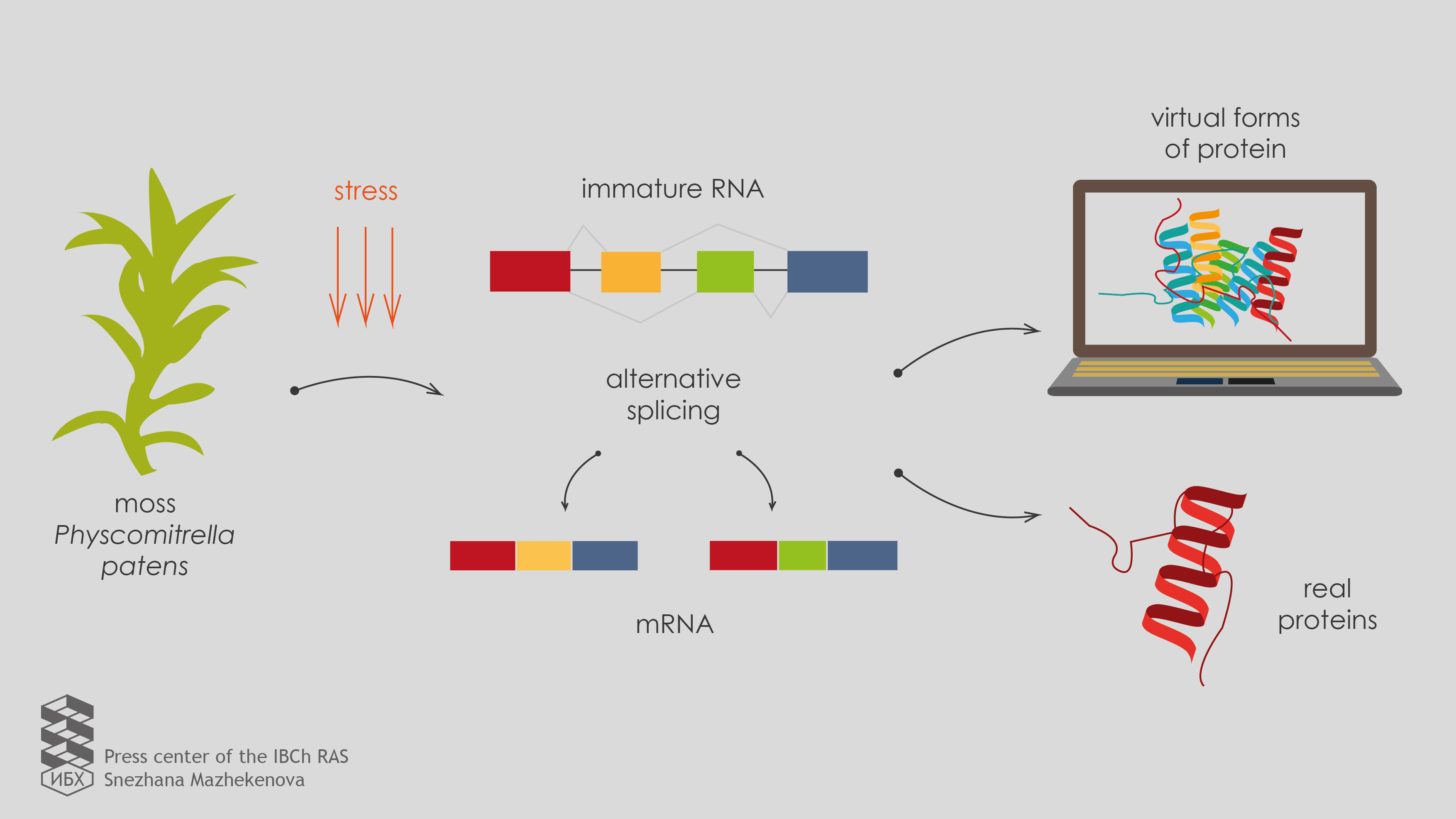
Eukaryotic messenger RNAs (mRNAs) can be undergone alternative splicing – the process that can generate several mRNAs from one gene. Thus, it was previously assumed that one gene can yield several mRNAs and, as a consequence, several protein isoforms.
"When scientists found that eukaryotic genes could undergo alternative splicing, it was suggested that due to this process, one gene can encode a huge number of diverse proteins. Partially this is true as some genes indeed produce different protein isoforms i. But in our study we showed that this is more exception than a rule. Earlier, studying the role of alternative splicing in a variety of processes in plants, biologists have not taken this moment into account in their work," says Igor.
Author: Snezhana Mazhekenova.
Igor and his colleagues began their research with standard methods for the analysis of gene expression. They extracted RNA from different cell types of the model object (moss Physcomitrella patens), determined the primary structure (sequencing) and predicted protein isoforms which can be translated from such RNAs. Then the scientists treated the moss cells with various stress factors, repeated all the manipulations and found that the alternative splicing actively changes messenger RNAs. To confirm that predicted proteins are present in the cell and to check whether alternative splicing influences proteome diversity, scientists isolated proteins from moss cells and performed the analysis on a mass spectrometer. It turned out that the role of alternative splicing in the formation of the proteome composition of the cell is not so significant as it was expected.
“The reviewers of the journal, where we submitted the article, suggested that the results we obtained are associated with general limitations of mass spectrometry analysis. To prove our point, we modeled an experiment in which we estimated the number of isoforms that we had to identify if they were present in the cell. It turned out that if alternative splicing plays an important role in the protein composition formation in the cell, we would see dozens more isoforms in our mass spectrometric experimental data than we actually obtained.”
Now scientists have to understand biological function of alternative splicing and the role of RNA-RNA interactions in this process as well as this affects the translation of proteins in the cell.
august 16, 2017

