Press-room / news / Science news /
In silico prediction of transmembrane dimers for bitopic proteins
Laboratory of biomolecular modeling at Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry (IBCh) for several years has been developing computational technique for theoretical assessment of dimerization modes for transmembrane (TM) domains of receptor tyrosine kinases. This dimerization represents an important molecular event in cell growth and differentiation. A (rather) simple prediction tool for possible configuration of TM-dimers is now available as a PREDDIMER web service: http://model.nmr.ru/preddimer.
Transmembrane (TM) domains of bitopic proteins, such as receptor tyrosine kinases (RTKs), comprise just single α-helix. These receptors are involved in cell division and growth and sense various growth factors and insulin. A receptor of alkaline media (homologous to insulin receptor) that has been puzzled out at IBCh several years ago [1], also belongs to RTK family. It has been shown for RTKs that TM-domains play a crucial role in dimerization and activation of these receptors. The function of TM-helices — signal transmitters between extracellular and intracellular RTK parts — is modulation of mutual orientation of intracellular kinase domains and subsequent control of self-phosphorylation, which represents the initial input into the signal amplification cascade. Even single-point mutations in RTK TM-domains may cause a number of severe disorders including human inherited diseases and cancer. Despite large experimental effort, the mechanism of such effects is still under debates.
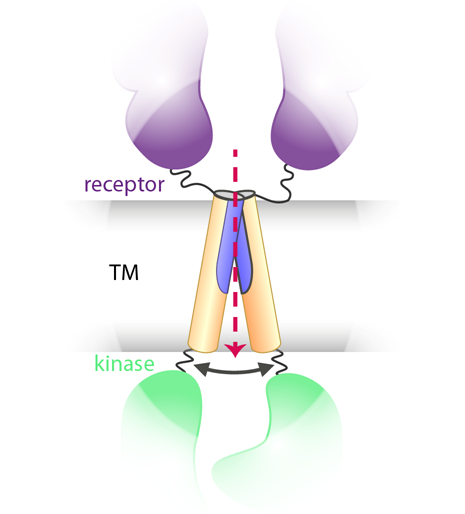
Transmembrane (TM) helices of receptor tyrosine kinases are signal transmitters between extracellular (receptor domains) and intracellular (kinase domains) protein fragments. They modulate the mutual orientation of intracellular kinase domains and subsequently control self-phosphorylation of the receptors.
Although a certain progress in experimental structure determination of TM-dimers by NMR spectroscopy has been recently achieved (mainly, by the Laboratory of Biomolecular NMR spectroscopy headed by Professor Alexander Arseniev), current limitations of the existing techniques still make no clear answer of how biologically reliable are the obtained dimer conformations and whether any conformations alternative to NMR model may be adopted by TM-dimers upon protein functioning as well as in response to changes of the local lipid environment.
Molecular modeling represents a reasonable alternative way to understand structural organization of TM helical dimers. The existing approaches vary a lot in their complexity, prediction power and computational efficiency starting from simple sequence motif-based algorithms to laborious force field-based techniques like Monte-Carlo conformational search in the implicit membrane, coarse-grained or all atom molecular dynamics simulations in the explicit lipid bilayers, or different combinations of them. In a number of recent studies carried out in the Laboratory of Biomolecular Modeling headed by Professor Roman Efremov, these modeling approaches have been successfully applied to understanding conformational heterogeneity and thermodynamics of various RTKs’ TM-dimers [2]. It has been shown that RTK TM-dimers are indeed able to adopt different conformations, whose membrane geometry and/or relative stability are governed and fine-tuned by the particular lipid environment [2, 3]. At the same time, pathogenic single-point mutations in TM-regions can induce conformational rearrangements of TM-dimers affecting their association strength in different states and lead to repopulation of their conformational ensembles, which may modulate RTK activation [4, 5].
In the context of prediction of heterogeneous conformational ensembles for TM-dimers, the Laboratory of Biomolecular Modeling implemented PREDDIMER web-server (http://model.nmr.ru/preddimer), which has been announced very recently in Bioinformatics [6]. The server allows robust reconstruction of a number of dimer structures for given sequences of TM-protein fragments and visualization of predicted dimers as 3D-models or 2D-maps of helical surface hydrophobicity with outlined contacting regions. The benchmark of the server on 11 TM-sequences, whose 3D dimer conformations were obtained previously by NMR spectroscopy, shows that in the most tested cases the algorithm is able to predict the experimental structures with backbone root-mean-square deviations from the reference below 3 Å.
Despite obvious limitation of PREDDIMER, which operates with ideal helices without explicit consideration of the membrane environment and outputs rather “coarse-grained” dimer configurations, the prediction results represent a reliable starting point for further more detailed and realistic modeling and/or give an opportunity for fast screening of dimerization possibilities of given TM-sequences.
References
- Press-release at IBCh website, 10.06.2011: “Receptor with ‘non-traditional’ orientation”;
-
Polyansky A.A., Volynsky P.E., Efremov R.G. (2012). Multistate organization of transmembrane helical protein dimers governed by the host membrane. J. Am. Chem. Soc. 134,
14390–143400; -
Muhle-Goll C., Hoffmann S., Afonin S., Grage S.L, Polyansky A.A., Windisch D., Zeitler M., Buerck J., Ulrich A.S. (2012). Hydrophobic matching controls the tilt and stability of the dimeric platelet-derived growth factor receptor (PDGFR) β transmembrane segment. J. Biol. Chem. 287,
26178–26186; - Velghe A.I., Van Cauwenberghe S., Polyansky A.A., Chand D., Montano-Almendras C.P., Charni S., Hallberg B., Essaghir A., Demoulin J.B. (2013). PDGFRA alterations in cancer: characterization of a gain-of-function V536E transmembrane mutant as well as loss-of-function and passenger mutations. Oncogene, doi:10.1038/onc.2013.218;
-
Volynsky P. E., Polyansky A. A., Fakhrutdinova G. N., Bocharov E. V., Efremov R. G. (2013). Role of dimerization efficiency of transmembrane domains in activation of fibroblast growth factor receptor 3. J. Am. Chem. Soc. 135,
8105–8108; - Polyansky A.A., Chugunov A.O., Volynsky P.E., Krylov N.A., Nolde D.E., Efremov R.G. (2013). PREDDIMER: a web server for prediction of transmembrane helical dimers. Bioinformatics, doi: 10.1093/bioinformatics/btt645.
december 24, 2013

